Monday, March 31, 2008
Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein.
http://www.doaj.org/doaj?func=abstract&id=121871
History of DNA research
- Further information: History of molecular biology
DNA was first isolated by the Swiss physician Friedrich Miescher who, in 1869, discovered a microscopic substance in the pus of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein".[117] In 1919 this discovery was followed by Phoebus Levene's identification of the base, sugar and phosphate nucleotide unit.[118] Levene suggested that DNA consisted of a string of nucleotide units linked together through the phosphate groups. However, Levene thought the chain was short and the bases repeated in a fixed order. In 1937 William Astbury produced the first X-ray diffraction patterns that showed that DNA had a regular structure.[119]
In 1928, Frederick Griffith discovered that traits of the "smooth" form of the Pneumococcus could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form.[120] This system provided the first clear suggestion that DNA carried genetic information, when Oswald Avery, along with coworkers Colin MacLeod and Maclyn McCarty, identified DNA as the transforming principle in 1943.[121] DNA's role in heredity was confirmed in 1952, when Alfred Hershey and Martha Chase in the Hershey-Chase experiment showed that DNA is the genetic material of the T2 phage.[122]
In 1953, based on X-ray diffraction images[123] taken by Rosalind Franklin and the information that the bases were paired, James D. Watson and Francis Crick suggested[123] what is now accepted as the first accurate model of DNA structure in the journal Nature.[5] Experimental evidence for Watson and Crick's model were published in a series of five articles in the same issue of Nature.[124] Of these, Franklin and Raymond Gosling's paper was the first publication of X-ray diffraction data that supported the Watson and Crick model,[125][126] this issue also contained an article on DNA structure by Maurice Wilkins and his colleagues.[127] In 1962, after Franklin's death, Watson, Crick, and Wilkins jointly received the Nobel Prize in Physiology or Medicine.[128] However, debate continues on who should receive credit for the discovery, as the Watson and Crick article in Nature was based on Franklin's data without either acknowledgment or her knowledge.[129]
In an influential presentation in 1957, Crick laid out the "Central Dogma" of molecular biology, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis".[130] Final confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 through the Meselson-Stahl experiment.[131] Further work by Crick and coworkers showed that the genetic code was based on non-overlapping triplets of bases, called codons, allowing Har Gobind Khorana, Robert W. Holley and Marshall Warren Nirenberg to decipher the genetic code.[132] These findings represent the birth of molecular biology.
http://en.wikipedia.org/wiki/DNANanotech C++
The enormous raw power of dna computing keeps the field moving in spite of all the daunting technical obstacles. Yet even if those obstacles ultimately prove insurmountable, Winfree's work could mean a breakthrough in the construction of ultrasmall devices. Indeed, Winfree himself thinks DNA tiles' most exciting application is as intelligent building blocks that put themselves together piece by piece on the nanometer scale-assembling into large and complex structures.
Collaborating with Rothemund and Adleman at USC, Winfree aims to fabricate a two-dimensional shape known as the Sierpinski triangle. Named after the Polish mathematician who discovered it in 1915, the triangle is a complex and beautiful fractal produced by repeating a simple geometric rule. The team plans to construct a real-world version of the triangle in a test tube using only seven different DNA tiles. Winfree has designed each tile type to carry out a simple program-to add itself to the growing shape or not, depending on the molecular cues provided by the triangle's outer edge.
In the hands of nanofabrication experts like NYU's Seeman, the DNA tiles could lead to easier methods to make exotic molecular structures-doing for nanotech what CAD and pre-fab building materials have done for the construction industry. "Greater control leads to things that you almost can't imagine," says Seeman. "Our expectation is that this approach can be applied to making designer materials and interesting patterns much more economically."
Seeman's lab, for instance, is already trying to attach nanoparticles of gold to DNA tiles in order to prototype tiny electrical circuits. These DNA "assemblies" would be about 10 times smaller than the tiniest features etched in silicon chips. However, Rothemund notes that there are limits to the patterns "computable" with DNA tiles. "We can't make anything we want," says Rothemund. "But the simple assemblies we've made so far show how well the basic operations work."
They also show how much scientists still have to learn. Winfree compares his efforts so far to one-line programs written in biochemical Basic. What he'd really like to be doing is programming biochemical reactions in C++. He expects this more advanced language will evolve as researchers master new operations, such as selectively removing tiles from an assembly. Winfree speculates that one day it may be possible to bring this growing repertoire of programmable components together to build synthetic systems-call them "nanorobots" if you wish-capable of independently carrying out useful tasks. "The really interesting direction DNA computing is taking us is to see just how far we can learn to program biochemical reactions," says Winfree.
That may sound like futuristic hype, but researchers are already beginning to figure out ways to do it. At Lucent Technologies' Bell Labs, physicist Bernie Yurke, for one, is working with DNA in the hopes of assembling ultrasmall molecular motors. Yurke imagines that some day it might be possible to build a DNA motor that could walk across Winfree's DNA tiling constructs, making chemical changes at specific points. "You could lay down an arbitrarily complex pattern," Yurke says, which might then be transferred to a silicon substrate to fabricate nanometer-scale circuits and transistors. "My hope is that in the future complicated electronic structures like computers will be made this way."
Electronic computers assembled using DNA that computes? It may sound like an unlikely twist in the evolution of DNA computing, but it's one that Adleman believes is entirely in keeping with the field he helped launch. "Like quantum computing, DNA computing is very futuristic, and both make the point that computation doesn't have to take place in the box that sits on our desks," says Adleman, this time in a telephone interview. "Even if they don't become a viable means of computing in the future-and I don't know if they will-we may learn what the real computer of the future should look like."
Computing (and Constructing) With DNA Organization Key Researchers Focus Bell Labs Bernie Yurke, Allan Mills Fabricating DNA motors for assembling electronic components Duke University/Caltech John Reif, Thomas LaBean, Erik Winfree (Caltech) Working on massively parallel addition using DNA tiles New York University Nadrian Seeman Assembling complex nanostructures out of DNA Princeton University Laura Landweber, Richard Lipton RNA-based computer used to solve chess puzzle known as the "knight problem" University of Southern California Leonard Adleman Automating a self-contained lab system for DNA computing; proved, in theory, that DNA can crack DES data encryption standard University of Wisconsin Robert M. Corn, Lloyd M. Smith, Anne E. Condon, Max G. Lagally Adapting DNA-chip technology to do DNA computation on a solid surface
http://www.technologyreview.com/Biotech/12108/page3/DNA Uses in technology
Genetic engineering
- Further information: Molecular biology and genetic engineering
Modern biology and biochemistry make intensive use of recombinant DNA technology. Recombinant DNA is a man-made DNA sequence that has been assembled from other DNA sequences. They can be transformed into organisms in the form of plasmids or in the appropriate format, by using a viral vector.[100] The genetically modified organisms produced can be used to produce products such as recombinant proteins, used in medical research,[101] or be grown in agriculture.[102][103]
Forensics
- Further information: Genetic fingerprinting
Forensic scientists can use DNA in blood, semen, skin, saliva or hair at a crime scene to identify a perpetrator. This process is called genetic fingerprinting, or more accurately, DNA profiling. In DNA profiling, the lengths of variable sections of repetitive DNA, such as short tandem repeats and minisatellites, are compared between people. This method is usually an extremely reliable technique for identifying a criminal.[104] However, identification can be complicated if the scene is contaminated with DNA from several people.[105] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[106] and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.[107] People convicted of certain types of crimes may be required to provide a sample of DNA for a database. This has helped investigators solve old cases where only a DNA sample was obtained from the scene. DNA profiling can also be used to identify victims of mass casualty incidents.[108]
Bioinformatics
- Further information: Bioinformatics
Bioinformatics involves the manipulation, searching, and data mining of DNA sequence data. The development of techniques to store and search DNA sequences have led to widely-applied advances in computer science, especially string searching algorithms, machine learning and database theory.[109] String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides.[110] In other applications such as text editors, even simple algorithms for this problem usually suffice, but DNA sequences cause these algorithms to exhibit near-worst-case behaviour due to their small number of distinct characters. The related problem of sequence alignment aims to identify homologous sequences and locate the specific mutations that make them distinct. These techniques, especially multiple sequence alignment, are used in studying phylogenetic relationships and protein function.[111] Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project, are difficult to use without annotations, which label the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by gene finding algorithms, which allow researchers to predict the presence of particular gene products in an organism even before they have been isolated experimentally.[112]
DNA nanotechnology
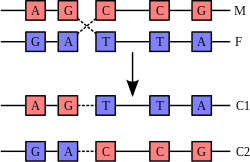
![The DNA structure at left (schematic shown) will self-assemble into the structure visualized by atomic force microscopy at right. DNA nanotechnology is the field which seeks to design nanoscale structures using the molecular recognition properties of DNA molecules. Image from Strong, 2004. [1]](http://upload.wikimedia.org/wikipedia/commons/thumb/5/55/DNA_nanostructures.png/400px-DNA_nanostructures.png)
- Further information: DNA nanotechnology
DNA nanotechnology uses the unique molecular recognition properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties. DNA is thus used as a structural material rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic lattices (both tile-based as well as using the "DNA origami" method) as well as three-dimensional structures in the shapes of polyhedra. Nanomechanical devices and algorithmic self-assembly have also been demonstrated, and these DNA structures have been used to template the arrangement of other molecules such as gold nanoparticles and streptavidin proteins.
History and anthropology
- Further information: Phylogenetics and Genetic genealogy
Because DNA collects mutations over time, which are then inherited, it contains historical information and by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny.[113] This field of phylogenetics is a powerful tool in evolutionary biology. If DNA sequences within a species are compared, population geneticists can learn the history of particular populations. This can be used in studies ranging from ecological genetics to anthropology; for example, DNA evidence is being used to try to identify the Ten Lost Tribes of Israel.[114][115]
DNA has also been used to look at modern family relationships, such as establishing family relationships between the descendants of Sally Hemings and Thomas Jefferson. This usage is closely related to the use of DNA in criminal investigations detailed above. Indeed, some criminal investigations have been solved when DNA from crime scenes has matched relatives of the guilty individual.[116]
http://en.wikipedia.org/wiki/DNADNA Dominoes
The idea of smart DNA tiles got its start five years ago at Caltech's Red Door cafe, when Winfree and Rothemund met to discuss Adleman's first DNA computing paper. The publication had set imaginations blazing throughout the world and across scientific disciplines. Were there other ways to compute with DNA? Could it beat silicon? Rothemund brought along a stack of papers showing "all the weirdest things that had been done with DNA." One of these was by Nadrian Seeman, a chemist at New York University who had created cubes, rings, octahedrons and other unlikely shapes from the DNA double helix. Winfree, who was working on a PhD related to artificial learning in robots, immediately saw a way that Seeman's strange versions of DNA could be used to compute.
Winfree's intellectual breakthrough was inspired by the theory of Wang tiles-a bit of recondite mathematics related to the patterns that can be created using squares with numbered sides. Like dominoes, the numbers on each Wang tile determine which other tiles it is allowed to touch. By carefully establishing these "matching rules," complex and interesting patterns can emerge as more tiles are added. But it's more than just a game of mathematical dominoes. Because Wang tiles carry both data (the numbers) and simple rules for combining it, mathematicians in the 1960s proved that the tiles could be used to add or multiply numbers. In fact, they showed that with the right set of these hypothetical constructs you could, in theory, do anything an electronic computer can-from playing chess to counting sheep. Winfree's big idea was a simple synthesis: use Seeman's DNA molecules as tiny real-life Wang tiles.
Applied to DNA computing, the strategy could sidestep one of the fundamental problems that has bedeviled the field from the beginning-too much lab work. While DNA computing is good at producing a vast number of answers quickly, things slow down when it comes to picking the right answers out of the mix. Take the traveling salesman problem originally solved by Adleman, in which the object is to find the most efficient route through seven cities connected by 14 one-way flights. Adleman created strands of DNA to represent each flight, then combined them in a test tube to generate every possible route.
Although the DNA in one-fiftieth of a teaspoon produced 100 trillion answers in less than one second, most of those answers were repeats-and most of them were incorrect. So Adleman's next task was to discard the wrong answers, something that could be done in a jiffy on a PC, but in Adleman's case required several dozen manual laboratory procedures. And that's where the trouble lies with most DNA computing schemes-each "operation" on the data means another time-consuming lab step.
The DNA tiles could solve that problem. Unlike the DNA used by Adleman in his original experiments that combined randomly, Winfree's tiles follow simple rules to get the correct result. "Ideally, you just put [the tiles] in the test tube and whammo!, you've got a right answer," says John Reif, a Duke University computer scientist.
Working with Winfree and Thom LaBean, a biochemist at Duke, Reif hopes to put the idea into practice by creating a simple molecular abacus out of DNA tiles. The goal is to add up binary numbers from zero to eight. With genetic letters standing in for 0s and 1s, the team has designed sets of tiles, each of which represents a possible column in an addition. Rules for combining columns correctly are coded into loose strands of DNA protruding from the sides of the tiles.
If all goes well, the experiment will generate several trillion multi-tile structures each of which has carried out an orderly addition of three binary bits. The scientists then will read off the results using standard methods for decoding DNA. The experiment underlines the potential power of DNA computers-massive parallelism and speed. Reif estimates that a single test tube of DNA tiles could perform about 10 trillion additions per second-about a million times faster than an electronic computer.
Evolution of DNA metabolism
- Further information: RNA world hypothesis
DNA contains the genetic information that allows all modern living things to function, grow and reproduce. However, it is unclear how long in the 4-billion-year history of life DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material.[81][93] RNA may have acted as the central part of early cell metabolism as it can both transmit genetic information and carry out catalysis as part of ribozymes.[94] This ancient RNA world where nucleic acid would have been used for both catalysis and genetics may have influenced the evolution of the current genetic code based on four nucleotide bases. This would occur since the number of unique bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribosomes.[95]
Unfortunately, there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible. This is because DNA will survive in the environment for less than one million years and slowly degrades into short fragments in solution.[96] Claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250-million years old,[97] but these claims are controversial.[98][99]
http://en.wikipedia.org/wiki/DNADNA Computing
Leonard Adleman sends his regrets. In an e-mail FAQ he uses to fend off journalists seeking interviews, the University of Southern California computer scientist and world-famous cryptographer who invented the field of DNA computing confesses that "http://www.technologyreview.com/Biotech/12108/are unlikely to become stand-alone competitors for electronic computers." He continues, somewhat apologetically: "We simply cannot, at this time, control molecules with the deftness that electrical engineers and physicists control electrons."
It was in 1994 that Adleman first used DNA, the molecule that our genes are made of, to solve a simple version of the "traveling salesman" problem. In this classic conundrum, the task is to find the most efficient path through several cities-given enough cities, the problem can challenge even a supercomputer. Adleman demonstrated that the billions of molecules in a drop of DNA contained raw computational power that might-just might-overwhelm silicon. But since then, scientists have run into tough practical and theoretical barriers. As Adleman and others in the field have come to realize, there may never be a computer made from DNA that directly rivals today'shttp://www.technologyreview.com/Biotech/12108/.
But that doesn't mean they've given up. Far from it. Although computer scientists haven't found a clear path from the test tube to the desktop, what they have found amazes and inspires them. Digital memory in the form of DNA and proteins. Exquisitely efficient editing machines that navigate through the cell, cutting and pasting molecular data into the stuff of life. What's more, nature packs all this molecular hi-fi equipment into a bacterium not much bigger than a single transistor. Viewed through the eyes of computer scientists, evolution has produced the smallest, most efficient computers in the world-and the beige-box set is hooked.
As Adleman now sees it, DNA computing is a field that's less about beating silicon than about surprising new combinations of biology and computer science that are pushing the limits in both fields-sometimes in unexpected directions. Scientists are still working hard on ways to tap the awesome number-crunching abilities of DNA for specialized types of applications, such as code breaking. But beyond that, the innate intelligence built into DNA molecules could help fabricate tiny, complex structures-in essence using computer logic not to crunch numbers but to build things.
Among the most promising of these new approaches are smart "DNA tiles" invented by Erik Winfree, a 30-year-old computer scientist at California Institute of Technology (see "100 Young Innovators," TR November/December 1999). Winfree's brainstorm is to create nanoscopic building blocks out of DNA that not only can store data but are designed-Winfree likes to say "programmed"-to carry out mathematical operations by fitting together in specific ways. Normally, DNA exists as two intertwined strands of the chemical letters A, G, C and T-the familiar double helix. But Winfree's DNA tiles are made by knotting together three or more of these strands, forming "tiles" about 15 nanometers (billionths of a meter) on their longest side. Taking advantage of DNA's ability to selectively recognize other strands of DNA, Winfree has "coded" the edges of these tiles so that they come together in just the right way to form tiny built-to-order structures.
In fact, programming DNA in this way could give chemists the kind of deft control "that may allow them to build more complex structures than any considered so far," says Paul Rothemund, a doctoral student in Adleman's USC lab.
Genetic recombination
- Further information: Genetic recombination
A DNA helix usually does not interact with other segments of DNA, and in human cells the different chromosomes even occupy separate areas in the nucleus called "chromosome territories".[87] This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is during chromosomal crossover when they recombine. Chromosomal crossover is when two DNA helices break, swap a section and then rejoin.
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection and can be important in the rapid evolution of new proteins.[88] Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.[89]
The most common form of chromosomal crossover is homologous recombination, where the two chromosomes involved share very similar sequences. Non-homologous recombination can be damaging to cells, as it can produce chromosomal translocations and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinases, such as RAD51.[90] The first step in recombination is a double-stranded break either caused by an endonuclease or damage to the DNA.[91] A series of steps catalyzed in part by the recombinase then leads to joining of the two helices by at least one Holliday junction, in which a segment of a single strand in each helix is annealed to the complementary strand in the other helix. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA.[92]
 |
 |
http://en.wikipedia.org/wiki/DNA
determining the structure of DNA
By the start of the 20th century, Cambridge stood out as one of the world's leading centers for science, of the same rank as the best German universities--Heidelberg, Göttingen, Berlin, and Munich. Over the next 50 years, Cambridge would remain in that rarefied league, but Germany would be supplanted by the United States, much strengthened by its absorption of many of the better Jewish scientists forced to flee Hitler. England similarly benefited from the arrival of some extraordinary Jewish intellectuals. If Max Perutz had not had the good sense to leave Austria in 1936 as a young chemist, there would have been no reason for my now moving to the banks of the Cam.
Though winning the great struggle against Hitler had drained England financially, the country's intellectuals took pleasure in knowing that victory had been much of their own making. Without the physicists who provided radar for British aviators during the Battle of Britain, or the Enigma code breakers of Bletchley Park who successfully pinpointed the German U-boats assaulting the Allies' Atlantic convoys, things might have turned out very differently.
Emboldened by the war to think expansively, the then tiny Medical Research Council (MRC) Unit for the Study of Structure of Biological Systems was doing science in the early 1950s that most chemists and biologists thought ahead of its time. Using x-ray crystallography to establish the 3-D structure of proteins was likely to be orders of magnitude more difficult than solving the structures of small molecules like penicillin. Proteins were daunting objectives, not only because of size and irregularity but because the sequence of the amino acids along their polypeptide chains was still unknown. This obstacle, however, was likely soon to be overcome. The biochemist Fred Sanger, working less than half a mile away from Max Perutz and John Kendrew at the MRC lab, was far along the path to establishing the amino acid sequences of the two insulin polypeptides. Others following in his steps would soon be working out the amino acid sequences of many other proteins.
https://www.technologyreview.com/Biotech/19173/page2/
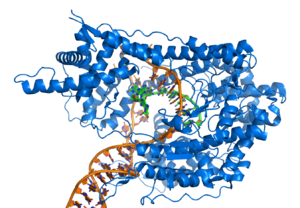
DNA-binding proteins
DNA-binding proteins
 |
 |
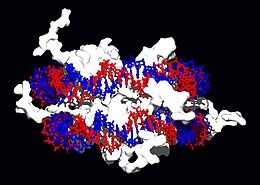
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called chromatin. In eukaryotes this structure involves DNA binding to a complex of small basic proteins called histones, while in prokaryotes multiple types of proteins are involved.[63][64] The histones form a disk-shaped complex called a nucleosome, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.[65] Chemical modifications of these basic amino acid residues include methylation, phosphorylation and acetylation.[66] These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factors and changing the rate of transcription.[67] Other non-specific DNA-binding proteins found in chromatin include the high-mobility group proteins, which bind preferentially to bent or distorted DNA.[68] These proteins are important in bending arrays of nucleosomes and arranging them into more complex chromatin structures.[69]
A distinct group of DNA-binding proteins are the single-stranded-DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication protein A is the best-characterised member of this family and is essential for most processes where the double helix is separated, including DNA replication, recombination and DNA repair.[70] These binding proteins seem to stabilize single-stranded DNA and protect it from forming stem-loops or being degraded by nucleases.
In contrast, other proteins have evolved to specifically bind particular DNA sequences. The most intensively studied of these are the various classes of transcription factors, which are proteins that regulate transcription. Each one of these proteins bind to one particular set of DNA sequences and thereby activates or inhibits the transcription of genes with these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[72] Alternatively, transcription factors can bind enzymes that modify the histones at the promoter; this will change the accessibility of the DNA template to the polymerase.[73]
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes.[74] Consequently, these proteins are often the targets of the signal transduction processes that mediate responses to environmental changes or cellular differentiation and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most accessible.[75]
DNA-modifying enzymes
Nucleases and ligases
Nucleases are enzymes that cut DNA strands by catalyzing the hydrolysis of the phosphodiester bonds. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut within strands. The most frequently-used nucleases in molecular biology are the restriction endonucleases, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5′-GAT|ATC-3′ and makes a cut at the vertical line. In nature, these enzymes protect bacteria against phage infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system.[77] In technology, these sequence-specific nucleases are used in molecular cloning and DNA fingerprinting.
Enzymes called DNA ligases can rejoin cut or broken DNA strands.[78] Ligases are particularly important in lagging strand DNA replication, as they join together the short segments of DNA produced at the replication fork into a complete copy of the DNA template. They are also used in DNA repair and genetic recombination.[78]
Topoisomerases and helicases
Topoisomerases are enzymes with both nuclease and ligase activity. These proteins change the amount of supercoiling in DNA. Some of these enzyme work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.[23] Other types of these enzymes are capable of cutting one DNA helix and then passing a second strand of DNA through this break, before rejoining the helix.[79] Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.[24]
Helicases are proteins that are a type of molecular motor. They use the chemical energy in nucleoside triphosphates, predominantly ATP, to break hydrogen bonds between bases and unwind the DNA double helix into single strands.[80] These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases
Polymerases are enzymes that synthesize polynucleotide chains from nucleoside triphosphates. The sequence of their products are copies of existing polynucleotide chains - which are called templates. These enzymes function by adding nucleotides onto the 3′ hydroxyl group of the previous nucleotide in a DNA strand. Consequently, all polymerases work in a 5′ to 3′ direction.[81] In the active site of these enzymes, the incoming nucleoside triphosphate base-pairs to the template: this allows polymerases to accurately synthesize the complementary strand of their template. Polymerases are classified according to the type of template that they use.
In DNA replication, a DNA-dependent DNA polymerase makes a DNA copy of a DNA sequence. Accuracy is vital in this process, so many of these polymerases have a proofreading activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to 5′ exonuclease activity is activated and the incorrect base removed.[82] In most organisms DNA polymerases function in a large complex called the replisome that contains multiple accessory subunits, such as the DNA clamp or helicases.[83]
RNA-dependent DNA polymerases are a specialized class of polymerases that copy the sequence of an RNA strand into DNA. They include reverse transcriptase, which is a viral enzyme involved in the infection of cells by retroviruses, and telomerase, which is required for the replication of telomeres.[84][32] Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure.[33]
Transcription is carried out by a DNA-dependent RNA polymerase that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a messenger RNA transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases, RNA polymerase II, the enzyme that transcribes most of the genes in the human genome, operates as part of a large protein complex with multiple regulatory and accessory subunits.[85]
http://en.wikipedia.org/wiki/DNAWiring Up DNA
 |
| Hot-wired: By placing a double-stranded DNA segment in a gap in a single-walled carbon nanotube, researchers have measured the electrical properties of the biological molecule. Since even a single mismatch in the DNA letters affects the conductivity of the segment, the system could eventually be the basis of chemical sensors to detect mutations in DNA. Credit: Colin Nuckolls |
By wiring up DNA between two carbon nanotubes, researchers have measured the molecule's ability to conduct electricity. Introducing just a single letter change can drastically alter the DNA's resistance, the researchers found, a phenomenon that they plan to exploit with a device that can rapidly screen DNA for disease-linked mutations.
Measuring the electrical properties of DNA has proved tricky because the molecule and its attachments to electrodes tend to be very fragile. But in the new study, Colin Nuckolls, a professor of chemistry at Columbia University, in New York, teamed up with Jacqueline Barton, a professor of chemistry at Caltech, in Pasadena, CA, who's an expert in DNA charge transport. Nuckolls's group had previously developed a method for securely hooking up biological molecules to single-walled carbon nanotubes, which act as the electrodes in a miniscule circuit.
The researchers used an etching process to slice a gap in a carbon nanotube; they created a carboxylic acid group on the nanotube at each end of the gap. They then reacted these groups with DNA strands whose ends had been tagged with amine groups, creating tough chemical amide links that bond together the nanotubes and DNA. The amide linkages are robust enough to withstand enormous electrical fields.
The team estimated that DNA strands of around 15 base pairs (around 6 nanometers) in length had a resistance roughly equivalent to that of a similar-sized piece of graphite. This is a finding that the researchers might have expected since the chemical base pairs that constitute DNA create a stack of aromatic rings similar to those in graphite.
"In my opinion, the results of this work will survive, in contrast to many other publications on this topic," says chemist Bernd Giese, of the University of Basel, Switzerland. Previous estimates of DNA's conductivity have varied dramatically, Giese says, partly because it was unclear if the delicate DNA or its connection to electrodes had become damaged by the high voltages used. "One thinks one has burned the DNA to charcoal," Giese says. "It's extremely complicated experimentally."
Barton and Nuckolls performed two tricks with their wired-up DNA. For their first, they introduced a restriction enzyme that bound and cut the DNA at a specific sequence. When severed, the current running through the DNA vanished. "It's a way of biochemically blowing a fuse," Nuckolls says. It also demonstratesthat the DNA keeps its native structure in the circuit; if it had not, the enzyme would not recognize and cut the molecule.
For their second trick, the researchers introduced a single base-pair mismatch into the DNA so that, for example, a C was paired up with an A (rather than its normal partner, G). This tweak boosted the molecule's resistance some 300-fold, probably because it distorts the double helical structure. They could do this easily by connecting only one of DNA's two strands into the circuit. The second strand - which can either be a perfect match to the first or contain a mismatch - can lift on or off.
Showing the electrical effect of such sequence mismatch and enzyme cutting is the real strength of the experiments, says Danny Porath, of Hebrew University, in Jerusalem, Israel, who has also measured current through DNA. "They play with the parameters and show that conductivity of DNA clearly depends on them, and that's beautiful," he says.
Nuckolls is now working to exploit this discovery to detect single nucleotide polymorphisms (SNPs), the one-letter variations in DNA that are linked to, for example, susceptibility to Alzheimer's, diabetes, and many other major diseases. Nuckolls hopes that his method can be used to identify SNPs more rapidly and with greater sensitivity than existing methods. In such a device, a reference strand of DNA is wired into the circuit and other strands allowed to pair up with it. If the second strand carries a different base at the position of the SNP, this would be enough to trigger a change in the current through a nanoscale circuit, just as the base-pair mismatch did. Nuckolls says that he is already working with electrical engineers to create a sensor that can slot into existing semiconductor chips, making it cheap and readily available. "It's one of our big focuses, and we're pretty close," he says.
The team is likely to have competition. Late last year, a group led by Wonbong Choi, of Florida International University, in Miami, reported that it had strung 80 base pairs of DNA between two carbon nanotubes and sent current through the DNA. Choi says that he is working to create a sensor that can rapidly reveal the presence of specific genetic sequences--such as the avian influenza virus--by looking at changes in current through the tiny circuit.
Barton, meanwhile, is intent on finding out whether the conductivity of DNA serves any biological purpose in the cell. She has evidence that proteins bound to DNA may detect DNA damage by changes in its electrical properties, perhaps triggering repair of the damage. "We think it's something nature takes advantage of," she says. "It's a radical idea, but I think as we get more and more evidence, the case will be built."
http://www.technologyreview.com/Nanotech/20205/Genes and genomes
- Further information: Cell nucleus, Chromatin, Chromosome, Gene, Noncoding DNA
Genomic DNA is located in the cell nucleus of eukaryotes, as well as small amounts in mitochondria and chloroplasts. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid.[54] The genetic information in a genome is held within genes, and the complete set of this information in an organism is called its genotype. A gene is a unit of heredity and is a region of DNA that influences a particular characteristic in an organism. Genes contain an open reading frame that can be transcribed, as well as regulatory sequences such as promoters and enhancers, which control the transcription of the open reading frame.
In many species, only a small fraction of the total sequence of the genome encodes protein. For example, only about 1.5% of the human genome consists of protein-coding exons, with over 50% of human DNA consisting of non-coding repetitive sequences.[55] The reasons for the presence of so much non-coding DNA in eukaryotic genomes and the extraordinary differences in genome size, or C-value, among species represent a long-standing puzzle known as the "C-value enigma."[56] However, DNA sequences that do not code protein may still encode functional non-coding RNA molecules, which are involved in the regulation of gene expression.[57]
Some non-coding DNA sequences play structural roles in chromosomes. Telomeres and centromeres typically contain few genes, but are important for the function and stability of chromosomes.[33][59] An abundant form of non-coding DNA in humans are pseudogenes, which are copies of genes that have been disabled by mutation.[60] These sequences are usually just molecular fossils, although they can occasionally serve as raw genetic material for the creation of new genes through the process of gene duplication and divergence.[61]
Transcription and translation
- Further information: Genetic code, Transcription (genetics), Protein biosynthesis
A gene is a sequence of DNA that contains genetic information and can influence the phenotype of an organism. Within a gene, the sequence of bases along a DNA strand defines a messenger RNA sequence, which then defines one or more protein sequences. The relationship between the nucleotide sequences of genes and the amino-acid sequences of proteins is determined by the rules of translation, known collectively as the genetic code. The genetic code consists of three-letter 'words' called codons formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
In transcription, the codons of a gene are copied into messenger RNA by RNA polymerase. This RNA copy is then decoded by a ribosome that reads the RNA sequence by base-pairing the messenger RNA to transfer RNA, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons (43 combinations). These encode the twenty standard amino acids, giving most amino acids more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAA, TGA and TAG codons.

Replication
- Further information: DNA replication
Cell division is essential for an organism to grow, but when a cell divides it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for DNA replication. Here, the two strands are separated and then each strand's complementary DNA sequence is recreated by an enzyme called DNA polymerase. This enzyme makes the complementary strand by finding the correct base through complementary base pairing, and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5′ to 3′ direction, different mechanisms are used to copy the antiparallel strands of the double helix.[62] In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
Interactions with proteins
All the functions of DNA depend on interactions with proteins. These protein interactions can be non-specific, or the protein can bind specifically to a single DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
http://en.wikipedia.org/wiki/DNAMolecular and biomolecular monolayers on diamond as an interface to biology
http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TWV-4DTTMB0-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=60543ee8ac4dfe091837fe2d3cc66231
DNA Chemical modifications
Chemical modifications
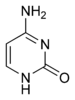 |  | 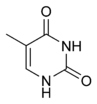 |
| cytosine | 5-methylcytosine | thymine |
Base modifications
- Further information: DNA methylation
The expression of genes is influenced by the chromatin structure of a chromosome and regions of that have low or no gene expression usually contain high levels of methylation of cytosine bases. For example, cytosine methylation, producing 5-methylcytosine, is important for X-chromosome inactivation.[38] The average level of methylation varies between organisms, with Caenorhabditis elegans lacking cytosine methylation, while vertebrates show higher levels, with up to 1% of their DNA containing 5-methylcytosine.[39] Despite the biological role of 5-methylcytosine it can deaminate to leave a thymine base, methylated cytosines are therefore particularly prone to mutations.[40] Other base modifications include adenine methylation in bacteria and the glycosylation of uracil to produce the "J-base" in kinetoplastids.[41][42]
DNA damage
- Further information: Mutation
DNA can be damaged by many different sorts of mutagens, which are agents that change the DNA sequence. These agents include oxidizing agents, alkylating agents and also high-energy electromagnetic radiation such as ultraviolet light and X-rays. The type of DNA damage produced depends on the type of mutagen. For example, UV light mostly damages DNA by producing thymine dimers, which are cross-links between adjacent pyrimidine bases in a DNA strand.[44] On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple forms of damage, including base modifications, particularly of guanosine, as well as double-strand breaks.[45] It has been estimated that in each human cell, about 500 bases suffer oxidative damage per day.[46][47] Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult to repair and can produce point mutations, insertions and deletions from the DNA sequence, as well as chromosomal translocations.[48]
Many mutagens intercalate into the space between two adjacent base pairs. Intercalators are mostly aromatic and planar molecules, and include ethidium, daunomycin, doxorubicin and thalidomide. In order for an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. These structural changes inhibit both transcription and DNA replication, causing toxicity and mutations. As a result, DNA intercalators are often carcinogens, with benzopyrene diol epoxide, acridines, aflatoxin and ethidium bromide being well-known examples.[49][50][51] Nevertheless, due to their properties of inhibiting DNA transcription and replication, they are also used in chemotherapy to inhibit rapidly-growing cancer cells.[52]
Overview of biological functions
DNA usually occurs as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. The set of chromosomes in a cell makes up its genome; the human genome has approximately 3 billion base pairs of DNA arranged into 46 chromosomes.[53] The information carried by DNA is held in the sequence of pieces of DNA called genes. Transmission of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA copy is then used to make a matching protein sequence in a process called translation which depends on the same interaction between RNA nucleotides. Alternatively, a cell may simply copy its genetic information in a process called DNA replication. The details of these functions are covered in other articles; here we focus on the interactions between DNA and other molecules that mediate the function of the genome.
http://en.wikipedia.org/wiki/DNA