Friday, March 14, 2008
Chromosome Structure and Function. Future Prospects
on “The Structure and Function of Chromatin”
that there has been a big advance in our understanding
of the three dimensional organization of
chromosomes at the first level of coiling of the DNA,
due mainly to the concept of the core nucleosome
particle (or ‘platysome’), although the exact location
of histone H1 and the arrangement of the linker
regions between platysomes are still in doubt. Even
less is known about the precise location of the nonhistone
proteins and the details of the higher levels of
coiling. Some 10 to 20% of the nucleosomes (the
amount depends on the tissue under study) have a
looser configuration which makes them more sensitive
to the nuclease DNase I. These ‘active’ nucleosomes
appear to include much of those stretches of DNA
which are being transcribed in any particular tissue.
In addition several lines of work suggest that
(as claimed for Escherichiu coli) the DNA in eukaryotes
is arranged in ‘domains’. The average size of these
domains is estimated, in very round terms, to be about
50000 base-pairs but the distribution of sizes about
the average is as yet unknown. Nor is the exact nature
and functional significance of these domains at all
clear though there is no lack of informed guesses
on this point. In particular one would like to know
whether the nucleosomes in a single domain are, at
any one time, all in the ‘active’ state or all in the
more compact inactive state or whether, on the other
hand, nucleosomes of both types occur at the same
time in a single domain.
http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1432-1033.1978.tb12061.x?cookieSet=1
Chromosomal aberrations
Chromosomal aberrations are disruptions in the normal chromosomal content of a cell, and are a major cause of genetic conditions in humans, such as Down syndrome. Some chromosome abnormalities do not cause disease in carriers, such as translocations, or chromosomal inversions, although they may lead to a higher chance of having a child with a chromosome disorder. Abnormal numbers of chromosomes or chromosome sets, aneuploidy, may be lethal or give rise to genetic disorders. Genetic counseling is offered for families that may carry a chromosome rearrangement.
The gain or loss of chromosome material can lead to a variety of genetic disorders. Human examples include:
- Cri du chat, which is caused by the deletion of part of the short arm of chromosome 5. "Cri du chat" means "cry of the cat" in French, and the condition was so-named because affected babies make high-pitched cries that sound like a cat. Affected individuals have wide-set eyes, a small head and jaw and are moderately to severely mentally retarded and very short.
- Wolf-Hirschhorn syndrome, which is caused by partial deletion of the short arm of chromosome 4. It is characterized by severe growth retardation and severe to profound mental retardation.
- Down's syndrome, usually is caused by an extra copy of chromosome 21 (trisomy 21). Characteristics include decreased muscle tone, asymmetrical skull, slanting eyes and mild to moderate mental retardation.
- Edwards syndrome, which is the second most common trisomy after Down syndrome. It is a trisomy of chromosome 18. Symptoms include mental and motor retardation and numerous congenital anomalies causing serious health problems. Ninety percent die in infancy; however, those who live past their first birthday usually are quite healthy thereafter. They have a characteristic hand appearance with clenched hands and overlapping fingers.
- Patau Syndrome, also called D-Syndrome or trisomy-13. Symptoms are somewhat similar to those of trisomy-18, but they do not have the characteristic hand shape.
- Idic15, abbreviation for Isodicentric 15 on chromosome 15; also called the following names due to various researches, but they all mean the same; IDIC(15), Inverted dupliction 15, extra Marker, Inv dup 15, partial tetrasomy 15
- Jacobsen syndrome, also called the terminal 11q deletion disorder.[1] This is a very rare disorder. Those affected have normal intelligence or mild mental retardation, with poor expressive language skills. Most have a bleeding disorder called Paris-Trousseau syndrome.
- Klinefelter's syndrome (XXY). Men with Klinefelter syndrome are usually sterile, and tend to have longer arms and legs and to be taller than their peers. Boys with the syndrome are often shy and quiet, and have a higher incidence of speech delay and dyslexia. During puberty, without testosterone treatment, some of them may develop gynecomastia.
- Turner syndrome (X instead of XX or XY). In Turner syndrome, female sexual characteristics are present but underdeveloped. People with Turner syndrome often have a short stature, low hairline, abnormal eye features and bone development and a "caved-in" appearance to the chest.
- XYY syndrome. XYY boys are usually taller than their siblings. Like XXY boys and XXX girls, they are somewhat more likely to have learning difficulties.
- Triple-X syndrome (XXX). XXX girls tend to be tall and thin. They have a higher incidence of dyslexia.
- Small supernumerary marker chromosome. This means there is an extra, abnormal chromosome. Features depend on the origin of the extra genetic material. Cat-eye syndrome and isodicentric chromosome 15 syndrome (or Idic15) are both caused by a supernumerary marker chromosome, as is Pallister-Killian syndrome.
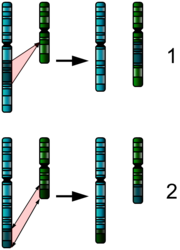
Chromosomal mutations produce changes in whole chromosomes (more than one gene) or in the number of chromosomes present.
- Deletion - loss of part of a chromosome
- Duplication - extra copies of a part of a chromosome
- Inversion - reverse the direction of a part of a chromosome
- Translocation - part of a chromosome breaks off and attaches to another chromosome
Most mutations are neutral - have little or no effect
A detailed graphical display of all human chromosomes and the diseases annotated at the correct spot may be found at [2].
Transmission of male infertility to future generations: lessons from the Y chromosome*
http://humupd.oxfordjournals.org/cgi/content/abstract/8/3/217
Karyotype
Although the replication and transcription of DNA is highly standardized in eukaryotes, the same cannot be said for their karotypes, which are often highly variable. There may be variation between species in chromosome number and in detailed organization. In some cases there is significant variation within species. Often there is variation 1. between the two sexes. 2. between the germ-line and soma (between gametes and the rest of the body). 3. between members of a population, due to balanced genetic polymorphism. 4. geographical variation between races. 5. mosaics or otherwise abnormal individuals. Finally, variation in karyotype may occur during development from the fertilised egg.
The technique of determining the karyotype is usually called karyotyping. Cells can be locked part-way through division (in metaphase) in vitro (in a reaction vial) with colchicine. These cells are then stained, photographed and arranged into a karyogram, with the set of chromosomes arranged, autosomes in order of length, and sex chromosomes (here XY) at the end: Fig. 3.
Like many sexually reproducing species, humans have special gonosomes (sex chromosomes, in contrast to autosomes). These are XX in females and XY in males.
Historical note
Investigation into the human karyotype took many years to settle the most basic question: how many chromosomes does a normal diploid human cell contain? In 1912, Hans von Winiwarter reported 47 chromosomes in spermatogonia and 48 in oogonia, concluding an XX/XO sex determination mechanism.[37] Painter in 1922 was not certain whether the diploid number of man was 46 or 48, at first favouring 46.[38] He revised his opinion later from 46 to 48, and he correctly insisted on man having an XX/XY system.[39] Considering their techniques, these results were quite remarkable.
New techniques were needed to definitively solve the problem:
- 1. Using cells in culture
- 2. Pretreating cells in a hypotonic solution, which swells them and spreads the chromosomes
- 3. Arresting mitosis in metaphase by a solution of colchicine
- 4. Squashing the preparation on the slide forcing the chromosomes into a single plane
- 5. Cutting up a photomicrograph and arranging the result into an indisputable karyogram.
It took until the mid 1950s until it became generally accepted that the karyotype of man included only 46 chromosomes.[40][41] Rather interestingly, chimpanzees (our closest living relatives) have 48 chromosomes.
The Chromosome Shuffle

Our genes are arrayed along 23 pairs of chromosomes. On rare occasion, a mutation can change their order. If we picture the genes on a chromosome as
ABCDEFGHIJKLMNOPQRSTUVWXYZ
a mutation might flip a segment of the chromosome, so that it now reads
ABCDEFGHISRQPONMLKJTUVWXYZ
or it might move one segment somewhere else like this:
ABCDLMNOPQRSTUEFGHIJKVWXYZ
In some cases, these changes can spread into the genome of an entire species, and be passed down to its descendant species. By comparing the genomes of other mammals to our own, scientists have discovered how the order of our genes has been shuffled over the past 100 million years. In tomorrow's New York Times I have an article on some of the latest research on this puzzle, focusing mainly on two recent papers you can read here and here.
One of the most interesting features of our chromosomes, which I mention briefly in the article, is that we're one pair short. In other words, we humans have 23 pairs of chromosomes, while other apes have 24. Creationists bring this discrepancy up a lot. They claim that it represents a fatal blow to evolution. Here's one account, from Apologetics Press:
If the blueprint of DNA locked inside the chromosomes codes for only 46 chromosomes, then how can evolution account for the loss of two entire chromosomes? The task of DNA is to continually reproduce itself. If we infer that this change in chromosome number occurred through evolution, then we are asserting that the DNA locked in the original number of chromosomes did not do its job correctly or efficiently. Considering that each chromosome carries a number of genes, losing chromosomes does not make sense physiologically, and probably would prove deadly for new species. No respectable biologist would suggest that by removing one (or more) chromosomes, a new species likely would be produced. To remove even one chromosome would potentially remove the DNA codes for millions of vital body factors. Eldon Gardner summed it up as follows: “Chromosome number is probably more constant, however, than any other single morphological characteristic that is available for species identification” (1968, p. 211). To put it another way, humans always have had 46 chromosomes, whereas chimps always have had 48.
There's a lot that's wrong here, and it can be summed up up with one number: 1968.
Why would someone quote from a 37-year-old genetics textbook in an article about the science of chromosomes? It's not as if scientists have been just sitting around their labs since then with their feet up on the benches. They've been working pretty hard, and they've learned a lot. And what they've learned doesn't agree with what Apologetics Press wants to claim.
The first big discovery came in 1982, when scientists looked at the patterns of bands on human and ape chromosomes. Chromosomes have a distinctive structure in their middle, called a centromere, and their tips are called telomeres. The scientists reported that the banding pattern surrounding the centromere on human chromosome 2 bore a striking resemblance to the telomeres at the ends of two separate chromosomes in chimpanzees and gorillas. They proposed that in the hominid lineage, the ancestral forms of those two chromosomes had fused together to produce one chromosome. The chromosomes weren't lost, just combined.
Other researchers followed up on this hypothesis with experiments of their own. In 1991, a team of scientists managed to sequence the genetic material in a small portion of the centromere region of chromosome 2. They found a distinctive stretches of DNA that is common in telomeres, supporting the fusion hypothesis. Since then, scientists have been able to study the chromosome in far more detail, and everything they've found supports the idea that the chromosomes fused. In this 2002 paper, for example, scientists at the Fred Hutchinson Cancer Research Center reported discovering duplicates of DNA from around the fusion site in other chromosomes. Millions of years before chromosome 2 was born, portions of the ancestral chromosomes were accidentally duplicated and then relocated to other places in the genome of our ancestors. And this past April, scientists published the entire sequence of chromosome 2 and were able to pinpoint the vestiges of the centromeres of the ancestral chromosomes--which are similar, as predicted, to the centromeres of the corresponding chromosomes in chimpanzees.
Today geneticists sometimes encounter people with fused chromosomes, which are often associated with serious disorders like Downs syndrome. But that doesn't mean that every fusion is harmful. Many perfectly healthy populations of house mice, for example, can be distinguished from other house mice by fused chromosomes. The fusion of chromosome 2 millions of years ago may not have caused any big change in hominid biology--except, perhaps, by making it difficult for populations of hominids with 23 pairs of chromosomes to mate with populations who still had 24. As a result, it may have helped produce a new species of hominid that would give rise to our own.
Just goes to show what 37 years of scientific research can turn up.
http://www.corante.com/loom/archives/2005/08/29/the_chromosome_shuffle.phpNumber of chromosomes in various organisms
Eukaryotes
These tables give the total number of chromosomes (including sex chromosomes) in a cell nucleus. For example, human cells are diploid and have 22 different types of autosomes, each present as two copies, and two sex chromosomes. This gives 46 chromosomes in total. Other organisms have more than two copies of their chromosomes, such as Bread wheat which is hexaploid and has six copies of 6 different chromosomes - 42 chromosomes in total.
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Normal members of a particular eukaryotic species all have the same number of nuclear chromosomes (see the table). Other eukaryotic chromosomes, i.e., mitochondrial and plasmid-like small chromosomes, are much more variable in number, and there may be thousands of copies per cell.  The 24 human chromosome territories during prometaphase in fibroblast cells. Asexually reproducing species have one set of chromosomes, which is the same in all body cells. Sexually reproducing species have somatic cells (body cells), which are diploid [2n] having two sets of chromosomes, one from the mother and one from the father. Gametes, reproductive cells, are haploid [n]: they have one set of chromosomes. Gametes are produced by meiosis of a diploid germ line cell. During meiosis, the matching chromosomes of father and mother can exchange small parts of themselves (crossover), and thus create new chromosomes that are not inherited solely from either parent. When a male and a female gamete merge (fertilization), a new diploid organism is formed. Some animal and plant species are polyploid [Xn]: they have more than two sets of homologous chromosomes. Agriculturally important plants such as tobacco or wheat are often polyploid compared to their ancestral species. Wheat has a haploid number of seven chromosomes, still seen in some cultivars as well as the wild progenitors. The more common pasta and bread wheats are polyploid, having 28 (tetraploid) and 42 (hexaploid) chromosomes compared to the 14 (diploid) chromosomes in the wild wheat.[34] ProkaryotesProkaryote species generally have one copy of each major chromosome, but most cells can easily survive with multiple copies.[35] Plasmids and plasmid-like small chromosomes are, like in eukaryotes, very variable in copy number. The number of plasmids in the cell is almost entirely determined by the rate of division of the plasmid - fast division causes high copy number, and vice versa. http://en.wikipedia.org/wiki/Chromosome | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Male chromosome may have a future after all
Researchers found no sign of gene loss over the past 6 million years, suggesting the chromosome is “doing a pretty good job of maintaining itself,” said researcher David Page of the Whitehead Institute for Biomedical Research in Cambridge, Mass.
That agrees with prior mathematical calculations that suggested the rate of gene loss would slow as the chromosome evolved, Page and study co-authors note in Thursday’s issue of the journal Nature. And, they say, it clashes with what Page called the “imminent demise” idea that says the Y chromosome is doomed to extinction.
The Y appeared 300 million years ago and has since eroded into a dinky chromosome, because it lacks the mechanism other chromosomes have to get rid of damaged DNA. So mutations have disabled hundreds of its original genes, causing them to be shed as useless. The Y now contains only 27 genes or families of virtually identical genes.In 2003, Page reported that the modern-day Y has an unusual mechanism to fix about half of its genes and protect them from disappearing. But he said some scientists disagreed with his conclusion. The new paper focuses on a region of the Y chromosome where genes can’t be fixed that way.
http://www.msnbc.msn.com/id/9146267/Chromosomes in prokaryotes
Structure in sequences
Prokaryotes chromosomes have less sequence-based structure than eukaryotes. Bacteria typically have a single point (the origin of replication) from which replication starts, while some archaea contain multiple replication origins.[17] The genes in prokaryotes are often organised in operons, and do not contain introns, unlike eukaryotes.
DNA packaging
Prokaryotes do not possess nuclei, instead their DNA is organized into a structure called the nucleoid.[18] The nucleoid is a distinct structure and occupies a defined region of the bacterial cell. This structure is however dynamic and is maintained and remodeled by the actions of a range of histone-like proteins, which associate with the bacterial chromosome.[19] In archaea, the DNA in chromosomes is even more organized, with the DNA packaged within structures similar to eukaryotic nucleosomes.[20][21]
Bacterial chromosomes tend to be tethered to the plasma membrane of the bacteria. In molecular biology application, this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes (and the attached DNA).
Prokaryotic chromosomes and plasmids are, like eukaryotic DNA, generally supercoiled. The DNA must first be released into its relaxed state for access for transcription, regulation, and replication.
http://en.wikipedia.org/wiki/ChromosomeThe History of Chromosomes May Shape the Future of Diseases
The common ancestor of humans and the rhesus macaque monkey lived about 25 million years ago. But despite that vast gulf of time, our chromosomes still retain plenty of evidence of our shared heritage.
A team of scientists at the National Cancer Institute recently documented this evidence by constructing a map of the rhesus macaque's DNA, noting the location of 802 genetic markers in its genome. Then they compared the macaque map to a corresponding map of the human genome. The order of thousands of genes was the same.
"About half of the chromosomes are pretty much intact," said William Murphy, a member of the team, now at Texas A&M University.
The other chromosomes had become rearranged over the past 25 million years, but Dr. Murphy and his colleagues were able to reconstruct their evolution. Periodically, a chunk of chromosome was accidentally sliced out of the genome, flipped around and inserted backward.
In other cases, the chunk was ferried to a different part of the chromosome. All told, 23 of these transformations took place, and within these blocks of DNA, the order of the genes remained intact.
"It's fairly easy to see how you can convert the chromosomes from the macaque to the human," Dr. Murphy said.
This new macaque study, which is set to appear in a future issue of the journal Genomics, is just one of many new papers charting the history of chromosomes - in humans and other species. While scientists have been studying chromosomes for nearly a century, only in the last few years have large genome databases, powerful computers and new mathematical methods allowed scientists to trace these evolutionary steps.
Scientists hope that uncovering the history of chromosomes will have practical applications to diseases like cancer, in which rearranged chromosomes play a major part.
Scientists have known for over 70 years that chromosomes can be rearranged. With a microscope, it is possible to make out the banded patterns on chromosomes and to compare the pattern in different species.
Scientists discovered that different populations of fruit fly species could be distinguished by inverted segments in their chromosomes.
Later, molecular biologists discovered how cells accidentally rearranged large chunks of genetic material as they made new copies of their chromosomes.
By the 1980's, scientists were able to identify some major events in chromosome evolution. Humans have 23 pairs of chromosomes, for example, while chimpanzees and other apes have 24. Scientists determined that two ancestral chromosomes fused together after the ancestors of humans split off from other apes some six million years ago.
But a more detailed understanding of how chromosomes had changed would have to wait until scientists had amassed more information. The mystery could not be solved with data alone. Deciphering the history of chromosomes is like a fiendishly difficult puzzle.
One well-studied version of it is known as the pancake problem. You have a stack of pancakes of different sizes, and you want to sort them into a neat pile from small to big. You can only do so by using a spatula to flip over some of the pancakes. Even a dozen pancakes make this a viciously hard problem to solve.
"Flipping chromosomes is a lot like flipping pancakes," said Pavel Pevzner of the University of California, San Diego.
In the mid-1990's, Dr. Pevzner and Sridhar Hannenhalli of the University of Pennsylvania invented a fast method for comparing chromosomes from two different species and determining the fewest number of rearrangements - the equivalent of pancake flips - that separate them.
They introduced the method with a series of talks with titles like "Transforming Cabbage Into Turnips" and "Transforming Mice Into Men."
"That opened the floodgates," said Bernard Moret of the University of New Mexico.
Scientists have used methods like Dr. Pevzner's to study different groups of species.
Dr. Pevzner himself joined with Dr. Murphy and 23 other scientists to analyze the last 100 million years of mammal evolution. They compared the genomes of humans to cats, dogs, mice, rats, pigs, cows and horses, using a program developed by Harris A. Lewin and his colleagues at the University of Illinois, called the Evolution Highway.
The program allowed them to trace how each lineage's chromosomes had become rearranged over time. They published their results in the July 22 issue of Science.
http://www.nytimes.com/2005/08/30/science/30gene.htmlChromosomes in eukaryotes
Eukaryotes (cells with nuclei such as plants, yeast, and animals) possess multiple large linear chromosomes contained in the cell's nucleus. Each chromosome has one centromere, with one or two arms projecting from the centromere, although under most circumstances these arms are not visible as such. In addition most eukaryotes have a small circular mitochondrial genome, and some eukaryotes may have additional small circular or linear cytoplasmic chromosomes.
In the nuclear chromosomes of eukaryotes, the uncondensed DNA exists in a semi-ordered structure, where it is wrapped around histones (structural proteins), forming a composite material called chromatin.
Chromatin
- Main article: Chromatin

Chromatin is the complex of DNA and protein found in the eukaryotic nucleus which packages chromosomes. The structure of chromatin varies significantly between different stages of the cell cycle, according to the requirements of the DNA.
[edit] Interphase chromatin
During interphase (the period of the cell cycle where the cell is not dividing) two types of chromatin can be distinguished:
- Euchromatin, which consists of DNA that is active, e.g., expressed as protein.
- Heterochromatin, which consists of mostly inactive DNA. It seems to serve structural purposes during the chromosomal stages. Heterochromatin can be further distinguished into two types:
- Constitutive heterochromatin, which is never expressed. It is located around the centromere and usually contains repetitive sequences.
- Facultative heterochromatin, which is sometimes expressed.
Individual chromosomes cannot be distinguished at this stage - they appear in the nucleus as a homogeneous tangled mix of DNA and protein.
Metaphase chromatin and division

In the early stages of mitosis or meiosis (cell division), the chromatin strands become more and more condensed. They cease to function as accessible genetic material (transcription stops) and become a compact transportable form. This compact form makes the individual chromosomes visible, and they form the classic four arm structure, a pair of sister chromatids attached to each other at the centromere. The shorter arms are called p arms (from the French petit, small) and the longer arms are called q arms (q follows p in the Latin alphabet). This is the only natural context in which individual chromosomes are visible with an optical microscope.
During divisions long microtubules attach to the centromere and the two opposite ends of the cell. The microtubules then pull the chromatids apart, so that each daughter cell inherits one set of chromatids. Once the cells have divided, the chromatids are uncoiled and can function again as chromatin. In spite of their appearance, chromosomes are structurally highly condensed which enables these giant DNA structures to be contained within a cell nucleus (Fig. 2).
The self assembled microtubules form the spindle, which attaches to chromosomes at specialized structures called kinetochores, one of which is present on each sister chromatid. A special DNA base sequence in the region of the kinetochores provides, along with special proteins, longer-lasting attachment in this region.
http://en.wikipedia.org/wiki/ChromosomePartitioning of Chromosomal DNA during Establishment of Cellular Asymmetry
How, then, are bacterial chromosomes prepared for asymmetrically positioned division events, such as that which occurs during the sporulation pathway of Bacillus subtilis? At the onset of sporulation, the chromosome is reorganized into a rod-like structure (the axial filament), which is readily distinguished from the bilobed chromosome of vegetative cells (31). Next, a septum is formed close to one cell pole, trapping the origin-proximal region of the chromosome in the smaller daughter cell (the forespore), while the remainder is translocated across the septum after division (43, 44). This translocation event requires the SpoIIIE protein (1, 43), without which the forespore receives only the origin-proximal 30% of a chromosome, while the remainder is located in the mother cell (44, 45). Thus, in the B. subtilis sporulation pathway, the relative order of division and chromosome segregation is reversed, with division occurring prior to chromosome segregation, rather than after.
Little is known about the architecture of the forespore chromosome before the sporulation septum is synthesized. Is the region of the invaginating septum cleared of DNA prior to asymmetric septation, as during vegetative growth, or is the septum required to partition the chromosome? Previous studies have supported the latter proposal, largely due to the failure to observe partitioned axial filaments in the absence of septation (45). The presence of a partitioned axial filament with a condensed region of the chromosome near one cell pole was therefore thought to indicate that a sporulation septum had formed (13, 21, 29, 45), implying that chromosome partitioning was a consequence of cell division. However, these studies preceded reliable methods to simultaneously observe the asymmetrically positioned sporulation septum and the chromosomes (27), leaving open the possibility that individual chromosomes are reorganized into two separate domains, creating a gap to accommodate the invaginating septum. Here, we address this issue using time-lapse deconvolution microscopy together with fluorescent membrane stains that allow visualization of septa and nucleic acid stains to reveal chromosome structure. We demonstrate that, during sporulation, the bacterial chromosome is partitioned into two domains of unequal size prior to asymmetric cell division. This asymmetric chromosome condensation event is independent of cell division protein FtsZ, providing evidence for an FtsZ-independent reorganization of cellular architecture that serves to prepare the bacterium for asymmetric cell division.
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=134875Visual discovery of chromosomes.
History of chromosomes
This is a brief history of research in a complex field where each advance was hard won, and often hotly disputed at the time.
Visual discovery of chromosomes. Textbooks have often said that chromosomes were first observed in plant cells by a Swiss botanist named Karl Wilhelm von Nägeli in 1842.[1] However, this opinion has been challenged, perhaps decisively, by Henry Harris, who has freshly reviewed the primary literature.[2] In his opinion the claim of Nägeli to have seen spore mother cells divide is mistaken, as are some of his interpretations. Harris considers other candidates, especially Wilhelm Hofmeister, whose publications in 1848-9 include plates which definitely show mitotic events.[3][4] Hofmeister was also the choice of Cyril Darlington.
The work of other cytologists such as Walther Flemming, Eduard Strasburger, Otto Bütschli, Oskar Hertwig and Carl Rabl should definitely be acknowledged. The use of basophilic aniline dyes was a new technique for effectively staining the chromatin material in the nucleus. Their behavior in animal (salamander) cells was later described in detail by Walther Flemming, who in 1882 "provided a superb summary of the state of the field".[5][6] The name chromosome was invented in 1888 by Heinrich von Waldeyer. However, van Beneden's monograph of 1883 on the fertilised eggs of the parasitic roundworm Ascaris megalocephala was the outstanding work of this period.[7] His conclusions are classic:
- Thus there is no fusion between the male chromatin and the female chromatin at any stage of division...
- The elements of male origin and those of female origin are never fused together in a cleavage nucleus, and perhaps they remain distinct in all the nuclei derived from them. [tranl: Harris p162]
"It is not easy to identify who first discerned chromosomes during mitosis, but there is no doubt that those who first saw them had no idea of their significance... [but] with the work of Balbiani and van Beneden we move away from... the mechanism of cell division to a precise delineation of chromosomes and what they do during the division of the cell." [8]
Van Beneden's master work was closely followed by that of Carl Rabl, who reached similar conclusions. [9] This more or less concludes the first period, in which chromosomes were visually sighted, and the morphological stages of mitosis were described. Coleman also gives a useful review of these discoveries.[10]
Nucleus as the seat of heredity. The origin of this epoch-making idea lies in a few sentences tucked away in Ernst Haeckel's Generelle Morphologie of 1866.[11] The evidence for this insight gradually acumulated until, after twenty or so years, two of the greatest in a line of great German scientists spelt it out. August Weismann proposed that the germ line was separate from the soma, and that the cell nucleus was the repository of the hereditary material, which he proposed was arranged along the chromosomes in a linear manner. Furthermore, he proposed that at fertilisation a new combination of chromosomes (and their hereditary material) would be formed. This was the explanation for the reduction division of meiosis (first described by van Beneden).
Chromosomes as vectors of heredity. In a series of outstanding experiments, Theodor Boveri gave the definitive demonstration that chromosomes were the vectors of heredity. His two principles were:
- The continuity of chromosomes
- The individuality of chromosomes.
It was the second of these principles which was so original. He was able to test the proposal put forward by Wilhelm Roux, that each chromosome carries a different genetic load, and showed that Roux was right. Upon the rediscovery of Mendel, Boveri was able to point out the connection between the rules of inheritance and the behaviour of the chromosomes. It is interesting to see that Boveri influenced two generations of American cytologists: Edmund Beecher Wilson, Walter Sutton and Theophilus Painter were all influenced by Boveri (Wilson and Painter actually worked with him). In his famous textbook The Cell, Wilson linked Boveri and Sutton together by the Boveri-Sutton theory. Mayr remarks that the theory was hotly contested by some famous geneticists: William Bateson, Wilhelm Johannsen, Richard Goldschmidt and T.H. Morgan, all of a rather dogmatic turn of mind. Eventually complete proof came from chromosome maps – in Morgan's own lab! [12]
http://en.wikipedia.org/wiki/ChromosomeUnderstanding Genetics

What Are Genetic Researchers Really Trying to Figure out?
Once researchers know which genes are involved in a disease, they can develop a test to screen people who are at risk and also start looking for a cure. A diagnostic test by itself will not cure the disease, but it can help identify high-risk people who may require more intensive screening or preventative action. Knowing what genes cause a given disease can also help researchers understand what goes wrong in that disease, which can help drive the search for drugs that counteract the problem
All Discoveries Are not of Equal ImportanceWhen newspapers announce that a gene has been discovered for a certain disease, it can really mean a number of things.
- Finding the gene: Sometimes, researchers have identified a gene that definitely causes a disease, such as the discovery of the gene for hemophilia or cystic fibrosis. Such a finding does not necessarily mean that researchers can cure the disease, or that a genetic test is immediately available, but it does mean that the medical community may be closer to an possible cure.
- Finding one of many genes: Other times, researchers have discovered a gene that plays a role in a small subset of people who get a common disease, such as the genes BRCA1 and BRCA2, which are the cause for some people's breast cancer. Again, finding these genes puts researchers one step closer to a cure or genetic test that can help some people with the disease.
- Finding a gene in animals: One way to understand gene function in in humans is to find and manipulate a gene that causes an animal — such as the mouse or fruit fly — to show symptoms similar to a human disease. Animals have genes that are very similar to our own, so these discoveries help point researchers to the biological function of a human disease gene. However, it is a long path from finding a gene in flies or mice to finding a genetic treatment for the human disease.
The Future Of Genetic Research
The eminent British molecular biologist Sydney Brenner once got a hearty laugh from his audience by describing how some future graduate student will define a mouse: "ATC, GCC, AAG, GGT, GTA, ATA. . . ." But every year the idea of defining an organism by the sequence of its DNA bases seems a little less farfetched.


In the sharpest image ever obtained of the DNA double helix (above,right) DNA is magnified approximately 25 million times with the aid of a scanning tunneling microscope, a powerful tool invented in the early 1980s. The turns and grooves of the DNA segment shown in this image closely match those of a corresponding computer-graphics model (above, left).
Victor McKusick, of The Johns Hopkins University School of Medicine, notes that scientists' growing ability to read and write in the language of the genes has already explained some of the once-mysterious basic concepts of genetics. The difference between dominant and recessive traits as causes of genetic disease used to be just an abstraction based on a great deal of observation. If a genetic defect expressed itself only in patients who inherited the trait from both parents, it was called recessive; both copies of the gene coding for the trait were presumably defective, resulting in disease. If the trait was dominant, on the other hand, it meant that one defective copy of the gene was sufficient to spell disaster.
But why should some disorders require two mistakes, while others resulted from only one? Molecular biology has given a concrete and remarkably simple explanation.
"It now appears that these two categories [recessive and dominant] correspond pretty closely to the two fundamental categories of proteins: enzymatic and structural," McKusick said in a recent review of genetics research. Recessive disorders tend to result from failures in genes that code for enzymes, the biological catalysts that do much of the body's chemical work. A person who has inherited the defective gene from only one parent often goes disease-free because the normal gene inherited from the other parent produces enough of the enzyme to serve the body's needs. The disorder appears only when the person inherits the same defect from both parents and therefore lacks any working copy of the normal gene.
If the genetic defect affects structural proteins, however, for example, collagen, a key component of connective tissues and bones, one copy of the faulty gene is usually enough to cause disease. It is easy to see why. A four-engine airplane can still fly even if one of its engines fails, as long as the other engines provide enough power, but a single faulty strut that makes a wing fall off will cause the plane to crash.
 The reason some genetic disorders are relatively common while most are extremely rare has also proved to be almost ridiculously obvious. The bigger the gene, the greater the chance that something will go wrong with part of it. In many cases, it seems as simple as that.
The reason some genetic disorders are relatively common while most are extremely rare has also proved to be almost ridiculously obvious. The bigger the gene, the greater the chance that something will go wrong with part of it. In many cases, it seems as simple as that.
Sometimes rather subtle differences in the defects of a single gene can make a profound difference in a patient's fate, as Louis Kunkel of the HHMI unit at Harvard University learned after he and his team discovered the gene for Duchenne muscular dystrophy (DMD) in 1986. Major flaws in that huge gene result in the presently incurable DMD, a muscle-wasting disease that leaves young boys wheelchair-bound by age 12 and generally kills them by age 20, because the muscles that control breathing fail. By contrast, lesser defects in that same gene produce a much more benign disease, Becker's muscular dystrophy.
A year after discovering this gene, the team identified the protein it codes for - a previously unknown protein, now named dystrophin, which occurs in muscles in such small amounts that it would never have been found by ordinary means. Dystrophin plays a key role in muscle cells and may be involved in many other muscle diseases. Researchers are now analyzing how dystrophin functions, what other proteins it interacts with, and whether it might be replaced to interrupt the course of disease.
Experts see many more insights such as these in the future, as research in molecular genetics opens some of the "black boxes" of biology.
"I think we are going to have an explosion of understanding," says David Valle, of the HHMI unit at The Johns Hopkins University. For example, the causes of mental disorders certainly include environmental factors, but biological psychiatrists believe the genes are whispering an important message, if only it can be heard.
Genetic research will illuminate many disorders of single organs, such as the eye, teeth, skin, and cochlea (the hearing apparatus of the ear), Valle believes. The deafness of about two-thirds of patients with serious hearing problems has a genetic basis, he says. Molecular biologists can find genes that are expressed only in the cochlea and therefore are probably important in hearing. Once such genes have been identified, several strategies exist for determining their functions and suggesting treatments.
Valle's current research focuses on a rare genetic disorder of the eye, gyrate atrophy, which leads to blindness through degeneration of the retina. The basic fault is an enzyme defect that causes an abnormal buildup of the amino acid ornithine. Surprisingly, some 35 different mutations in a single gene are able to produce the disease. The excess ornithine is found almost everywhere in the body - blood, urine, tears, spinal fluid, but the serious ill-effects are limited almost entirely to the retina. As yet, nobody knows why.
Understanding the genetic cause of the disease has led to a medical treatment that seems effective: severely restricting the patient's diet to bring the ornithine levels down to nearly normal. Recently, the scientists have compared the effects of this treatment on children in whom it was started early and on siblings who did not receive it until an older age. The studies confirm that the dietary restriction minimizes damage to the retina, Valle reports. But the diet is only a stopgap solution.
Geneticists are searching for more effective remedies, including possible treatment for the gene defect itself.
"One of the really exciting things about modern molecular genetics is that we now have opportunities to make animal models of these diseases and to study what happens at the tissue level in a direct way," Valle says. "That is one of the big things that is going to be happening in the next decade or so.
Philip Sharp, director of the Center for Cancer Research at MIT, divides the benefits of genetics research into two categories: those that generate knowledge and those that generate treatment. He sees animal models as extremely important to both. Deliberately produced genetic diseases in animals will have pathologies like those of human diseases. "We will learn how to recognize them, treat them, and analyze them in animals," he says. "That is going to be the forefront of biomedical science, in one area of it at least."
In addition, many aspects of human development will be clarified by work with mice, flies, and worms, he says. Scientists have discovered that genes which are developmentally active in both Drosophila, the fruit fly, and C. elegans, the nematode worm, have direct counterparts in mammals, although the functions of these genes in humans are not yet entirely clear.
When genes of species that separated from each other many millions of years ago show so much similarity, there is every reason to believe they are related. Many molecular biologists have noticed that nature is quite frugal in preserving devices that have proved biologically effective. As an example, Valle points out that the human enzyme ornithine delta aminotransferase, which is defective in gyrate atrophy, is 54 percent identical to the comparable enzyme that functions in yeast.
"I think one of the real themes of biology is that Mother Nature uses things over and over again once she figures out how to solve a problem," Valle says.
This concept offers scientists a great opportunity, says Eric Lander of the Whitehead Institute. He thinks there is hope of compiling, eventually, a complete thesaurus of protein parts that function in Earth's myriad species. "That would be spectacular," Lander says. "If we had the thesaurus of all the moving parts, then we would understand life in a remarkable way."
Gene mapping and cloning are key to the assembly of the thesaurus, and progress in these areas is clearly accelerating. However most of the 50,000 to 100,000 human genes remain totally unknown, and there is still a long way to go.
To date, most of the progress in understanding the genetics of human disease has involved relatively rare conditions, such as cystic fibrosis or Duchenne muscular dystrophy, which are caused by errors in single genes. But science is also stalking the genes that contribute to heart disease, cancer, diabetes, and mental illness - the big killers and cripplers of mankind.
It may soon be possible to tell some people that they have certain genetic predispositions to a specific major illness and suggest that they tailor their lifestyles accordingly. Similarly, the use of drugs to treat some of the important diseases could be tailored to the genetically varied needs of patients, with benefits for them and for the health care system in general: "Different strokes for different strokes," as one scientist put it.
On the other hand, some scientists fear that people might be stigmatized or become uninsurable because of genetic traits, such as carrier states, that don't in themselves have any appreciable effect on health.
Genetic research is advancing steadily, often rapidly, on many fronts. It has long been known that some disorders affect males, others affect females primarily, while still others may appear in either sex. But a few years ago researchers discovered that, even in some of the latter disorders the gravity and sometimes even the nature of disease may depend on which parent provided the faulty gene. This phenomenon is called imprinting. Although it has been detected only in rare human conditions, imprinting is a subject of intense study as researchers look for other examples.
Other scientists have forsaken the genes that reside in cell nuclei and are finding new clues to disease in the genes of what are probably our oldest and most entrenched "parasites" - the mitochondria - tiny, energy-generating organs inside every cell. Mitochondria are thought to be the descendants of ancient bacteria that not only found a home in animal cells, but also adapted so thoroughly that they became indispensable functional parts of those cells. We inherit mitochondria only from our mothers; sperm leave their mitochondria behind when they enter the egg. Flaws in mitochondrial genes have been found to lead to certain types of blindness and epilepsy and may also contribute to some degenerative disorders, such as dementia, which are associated with aging.
"Mitochondrial DNA gives us a whole new way to think about genetic transmission of diseases," says Douglas Wallace of Emory University, a specialist in those vital intracellular power stations.
In even more fundamental ways, discoveries in genetics have led to novel strategies for treating disease. Decades ago, scientists learned that DNA is mainly the archive of genetic information. Its orders are translated into action by segments of ribonucleic acid (RNA), which serve as the working blueprints for all proteins. Today, chemists are beginning to create valuable new drugs by fabricating "anti-sense" segments of RNA, whose sequence is the exact opposite of an unwanted sequence, to combine with certain existing strands of RNA and thus block the action of specific genes.
The bottom line in any kind of biomedical research lies in the realm of treatment and prevention. The ultimate step in that direction is gene therapy - the deliberate transplantation of genes to treat or even prevent human disease. Many geneticists dismiss gene therapy as a distant prospect, but others disagree. "We are going to have gene therapy," Philip Sharp says. "We are probably going to have it soon."
Gene therapy was actually tried in 1970 and again in 1980 without success, but the knowledge and techniques were primitive by today's standards. The first attempt in what might be called the modern era of gene therapy began in September 1990 at the National Institutes of Health (NIH), when doctors treated a 4-year-old girl. The child suffered from a grave immune deficiency because she lacked the enzyme adenosine deaminase. The doctors took her own white blood cells, altered them by adding the gene for the missing enzyme and transplanted the altered cells back into her.
Next on the NIH agenda was a substantially different strategy introducing a cancer-fighting substance, tumor necrosis factor, into the genetic repertoire of melanoma patients' own cancer-fighting white blood cells. Ultimately the same approach may be applied to other types of cancer.
Philip Sharp suggests one possibility that might be tried as soon as techniques are sufficiently refined. Instead of treating an AIDS patient for the rest of his or her life with a drug to protect the immune system against the HIV virus, doctors might use gene transplants to render the patient's immune system permanently resistant to the virus.
W. French Anderson of the NIH, one of the architects of the new attempts at gene therapy, sees a bright future. By the early years of the next century, he predicts, gene therapy will have become a highly sophisticated drug delivery system. Doctors will give the patient one, or perhaps several, transplants of his or her own cells that have been genetically engineered to manufacture a drug. In many cases this might replace the conventional practice of injecting drugs at regular intervals. How far in the future is this new application of genetic medicine? Five to ten years for the essential techniques, he estimates, somewhat longer to achieve a high degree of sophistication.
The first gene therapy attempts at NIH used the patient's white blood cells as the target for gene insertion. In the future, scientists hope to perfect techniques for using bone marrow cells. Several research centers are making progress in animal experiments using liver cells and endothelial cells, such as those that line blood vessels, to deliver valuable genes to the tissues where they would be useful. Another strategy that would have seemed sheer fantasy a few years ago is being discussed by serious scientists today. That is the idea of using inhalant spray to deliver copies of a good gene to airway tissues of cystic fibrosis patients.
The transplantation and manipulation of genes in other species has already proved valuable in genetics research and will probably play an even larger role in the future.
Mario Capecchi and his team at the University of Utah have recently used the method of gene manipulation known as homologous recombination to discover the function of a mouse gene. The gene first attracted notice because it produced breast cancer in the animals when it became activated abnormally. By developing mice in which that gene, and only that gene, had been knocked out, the scientists showed that the gene's normal function is crucial to the development of two regions of the animals' brain: the midbrain and cerebellum. The discovery opens an important door to studies of brain development and brain function.
To use homologous recombination, scientists must be able to identify and grow embryonic stem (ES) cells, the unspecialized precursors of all other cells in an organism. In Capecchi's mouse experiments, ES cells are modified to alter the specific gene under study and then implanted in a very early mouse embryo and used to breed animals that have the desired trait or flaw. Some experts consider this technique among the most exciting recent advances in genetics research.
But the excitement in genetics is general and pervasive. "Having been part of genetics research for 30 years, I find it almost stupefying that it is every bit as exciting and maybe even more so than it has seemed in the past," says Leon Rosenberg, dean of the Yale University School of Medicine. "I continue to be dazzled by the pace and surprise of new information in the field."
Studies of microbes, plants, animals, and many normal human beings are all contributing to the explosion of new knowledge. In recent years, molecular genetics has given important insights into the origin of life and its evolution, the emergence of humans, and our intimate relatedness to every other species on Earth. We can expect many more advances as geneticists continue to explore the wonder of life.